Great expectations have for the last few years been held for single-molecule technologies in DNA sequencing. So far, these expectations are far from being fulfilled. One expectation was that the read lengths should increase drastically, due to elimination of the problem of asynchrony, which limits read lengths in all multi-molecule methods. The combination of read length and accuracy seems to be difficult with single molecule sequencing, and investment cost has been at least as high with single molecule sequencing as with multimolecule variants. Nanopore based technologies have been held as the ultimate solution, but so far the basic problems with using nanopores have not been solved.
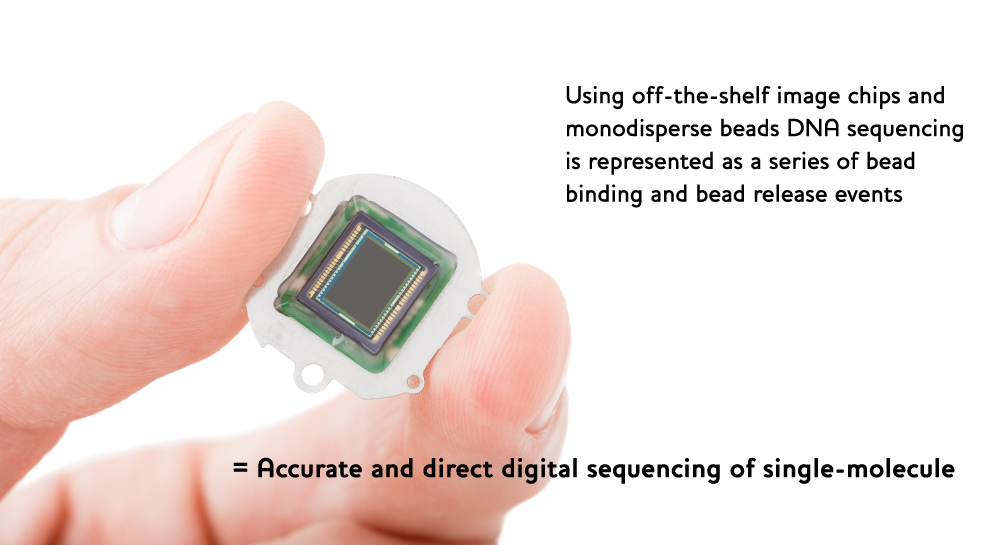
The Company´s MagSeq technology has the potential to combine low cost equipment and long read lengths of single molecule strands with high non-biased accuracy. It is based on the idea that DNA sequencing can be represented by a series of bead binding and bead release events.
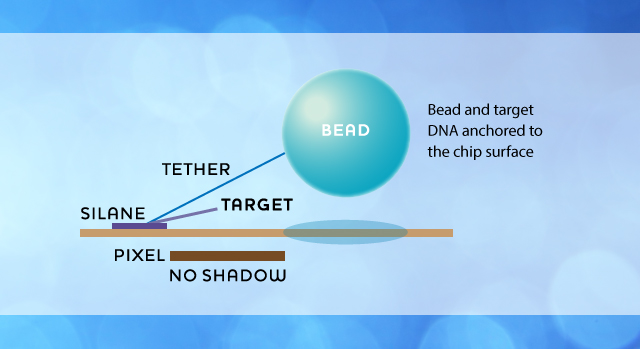
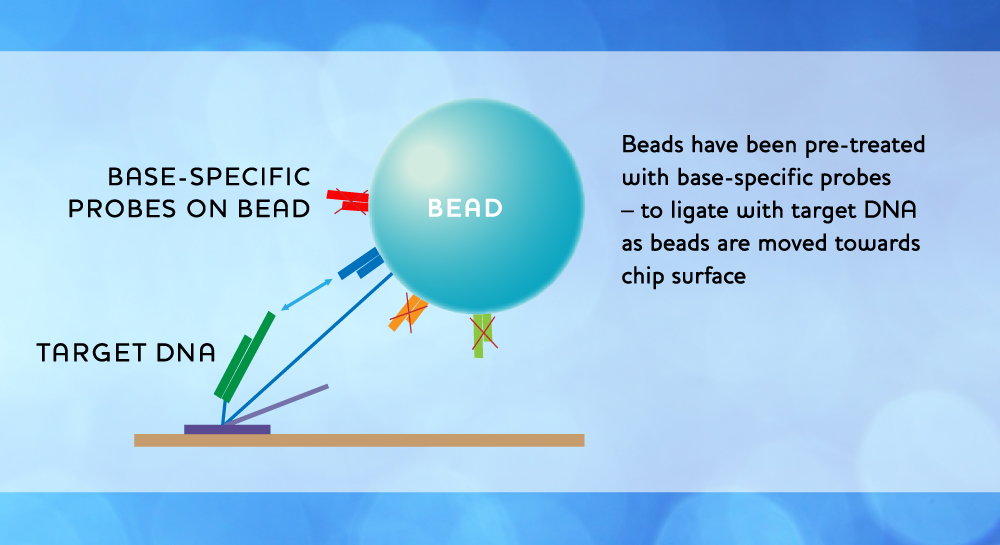
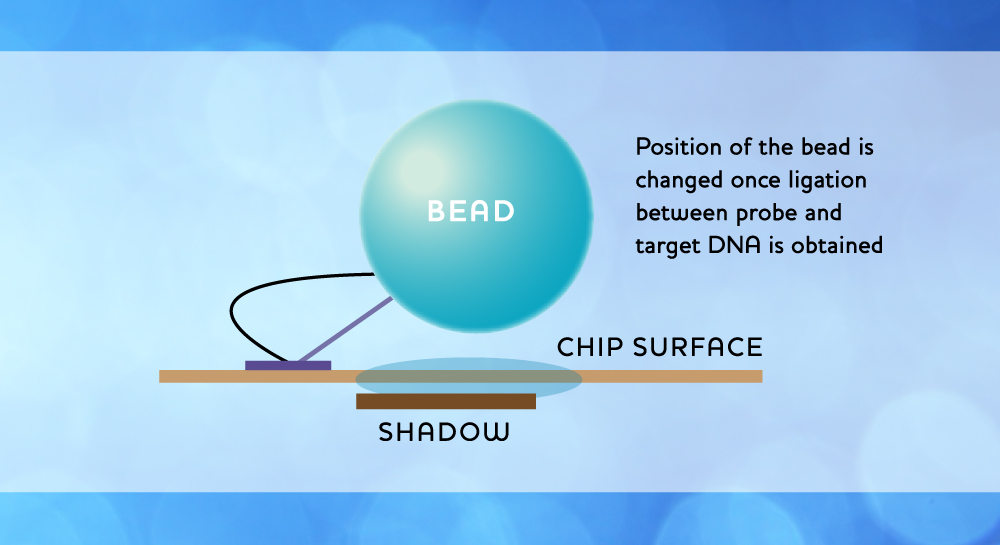
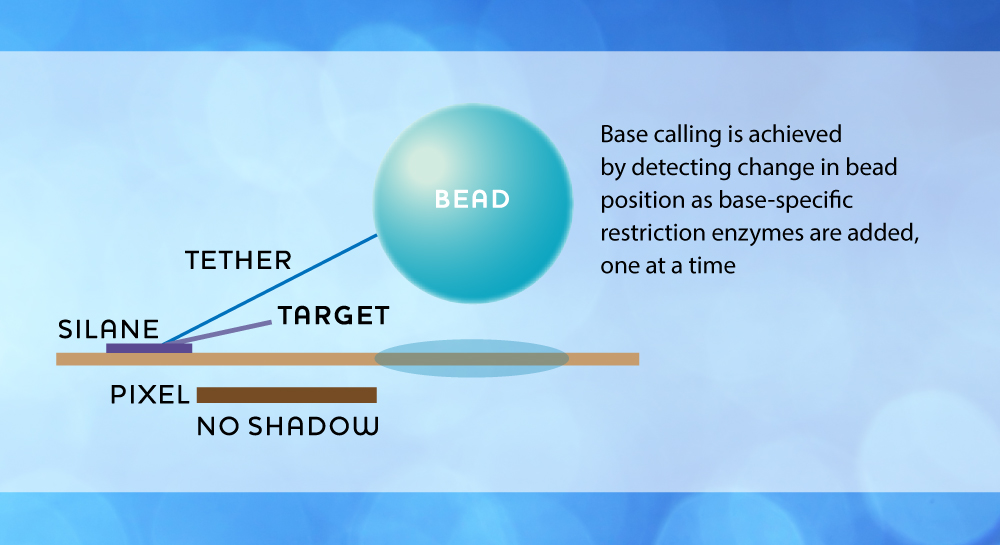
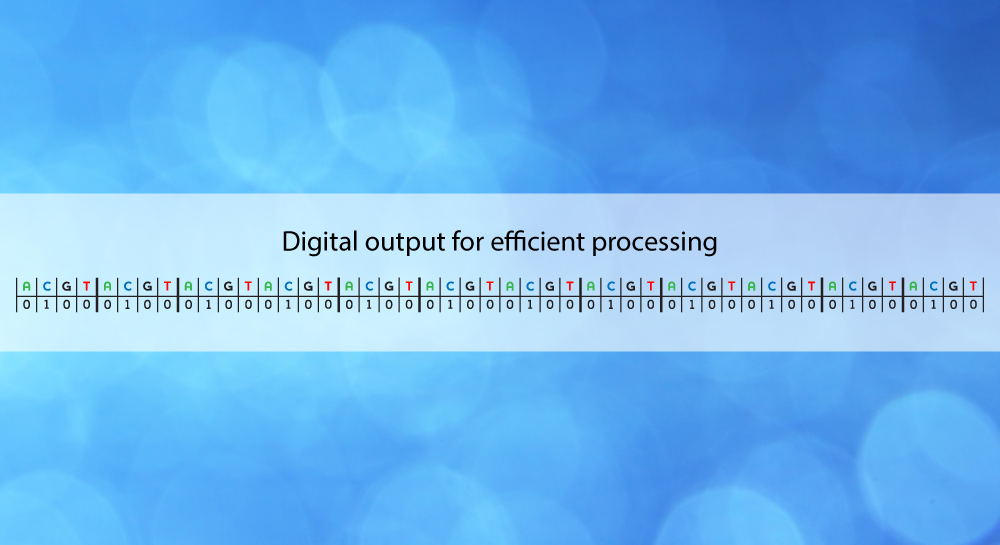
Using inexpensive and standard image sensors, the shadows of single beads immobilized on the surface of the chip are detected by simple off-the-shelf electronics. Base calling is extremely simple, as a pair of data bits, where each bit pair represents a bead/no-bead call. By using a look-up table, base calls can easily be translated into a DNA sequence string. Crucial in the chemical base recognition process are oligonucleotide probes, which by their base-specific binding to the DNA targets immobilize the beads. Probes are ligated to targets when probe and target overhangs match each other, and a cleavage reaction is used to expose the next base for reading and to reset the bead position. The process is controlled through sequential supply of fluids with registration of data by imaging for each supply cycle. There are four variants of supply systems, characterized by the way probes are supplied, i.e. either free or integrated with the beads, and by the way beads are supplied, i.e. through the fluid or permanently tethered to the chip surface. Depending upon the supply system variant used, the fluids will contain various combinations of enzymes, probes and beads. The MagSeq technology does not rely on any amplification steps, there is minimal labor involved, and the instrument and consumable costs are very moderate.
Status of Development
GeneSeque´s main focus has been to de-risk the key elements of the MagSeq technology concept. The company’s ambition is to fully implement the technology through collaboration with a development partner presently being sought. The development program to date has achieved the following de-risking milestones:
- Detection of beads directly on the surface of a standard image chip
- Development of technology to avoid non-specific bead binding on the chip surface
- Detection of beads immobilized on the surface through DNA anchoring
- Removal of beads through enzymatic release
- Base specific binding of probes on beads
- Construction of a suitable reaction chamber with appropriate fluid control
Several embodiments of the technology may be envisioned in order to achieve rapid and efficient sequencing. One possible solution is as illustrated below, where beads are kept relatively immobile around a pixel at all times through the use of an anchor molecule, while the sequencing reaction will be maintained by a DNA molecule connecting to a bead through base-specific binding.
Further work to be completed:
For full implementation of the preferred sequencing method, it remains to select chemistry compatible with the principles described above. With the suggested embodiment, beads will first be anchored on the pixels following a defined procedure. This procedure will eventually become a part of the prefabrication process.
Intellectual Property
GeneSeque has currently filed several patent applications in all major geographical patent territories: All the sequencing patents are in the national phase, while the sequencing length patent is in the PCT phase.